The diagnosis of IFDs is a multistep process.
It includes several core concepts:
Identifying patients
Diagnostic testing may be carried out to screen asymptomatic at-risk patients or patients with signs and symptoms.1 Whether or not diagnostic testing is carried out in patients may be confounded by the presence of nonspecific signs and symptoms.1
This can be of particular significance in patients in the intensive care unit (ICU) with multiple disease states, or in patients with haematologic malignancies and febrile neutropenia.1,2
Laboratory testing
The timeliness and accuracy of diagnostic testing are critical when an IFD is suspected, as delays in accurate diagnosis can lead to late initiation of therapy, and may be associated with poorer patient outcomes compared with early diagnosis.3
While conventional testing (culture testing, microscopy and histopathology) remains the ‘gold standard’ of laboratory testing for IFDs, limitations of these methods have led to the development of non-conventional methods, such as diagnostic biomarkers and polymerase chain reaction (PCR) assays.3,4
Complementary imaging techniques
Radiological imaging techniques, such as computed tomography, are often used to complement results from laboratory assays.5
Interpretation of results
Conventional diagnostic tests often require expert interpretation of results; however, recent developments in novel diagnostic assays have resulted in tests with simpler outputs, increasing the accessibility of IFD diagnosis.4

Diagnostic pathway
Use our interactive diagnostic pathway to discover the various diagnostic methods for IFDs
Guidelines
European and US guidelines are available to guide healthcare professionals in confirming an IFD diagnosis, including detail on different considerations in patient subgroups.6–9
The following three elements are used to optimise the accuracy of diagnosis, as the individual elements can be nonspecific in nature.7
Host factors
Such as recent history of neutropenia, haematological malignancy, receipt of a solid organ or allogenic stem cell transplantation or prolonged use of corticosteroids
Clinical features
Such as evidence of geographical or occupational exposure, acute localised pain, lesions, respiratory symptoms with cough, radiographic features or ophthalmological abnormalities
Mycological evidence
Such as positive diagnostic assay results (e.g. through culture, microscopic detection of fungal elements or molecular detection of fungal antigens)
Consensus definitions play a key role in fungal diagnostics
To standardise the definition of IFD, the European Organization for Research and Treatment of Cancer and the Mycoses Study Group Education and Research Consortium (EORTC/MSGERC) have developed definitions to classify IFDs into ‘proven’, ‘probable’ and ‘possible’. With the 2021 revision, the ‘probable’ definition has been expanded, while the scope of ‘possible’ has been reduced.10
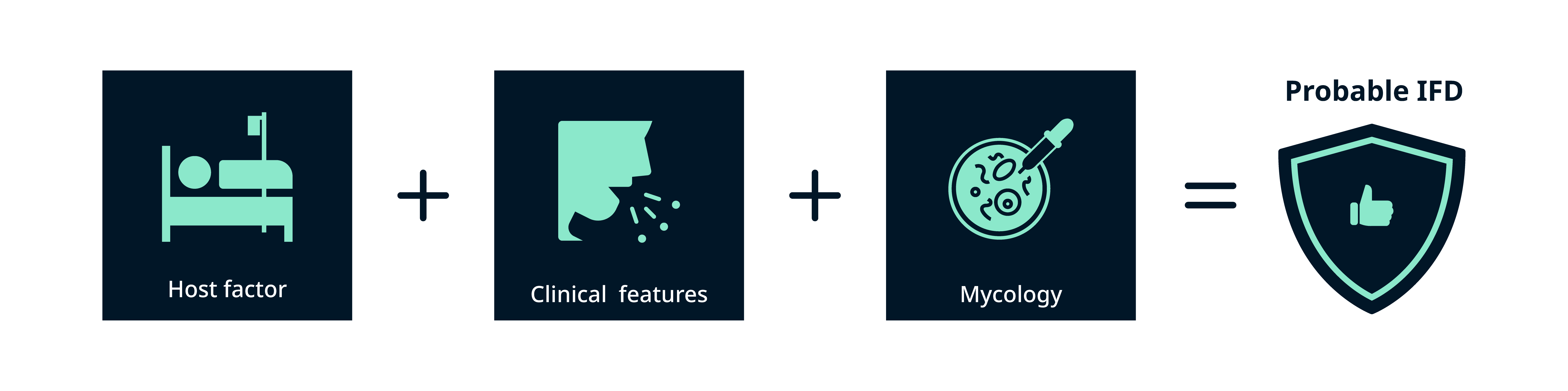
Probable IFD (proposed for immunocompromised patients) requires the presence of:7,10
- A host factor
- Clinical features
- Mycological evidence: culture and microscopy but also indirect tests
Possible IFD (proposed for immunocompromised patients) requires the presence of:7,10,11
- A host factor
- Clinical features

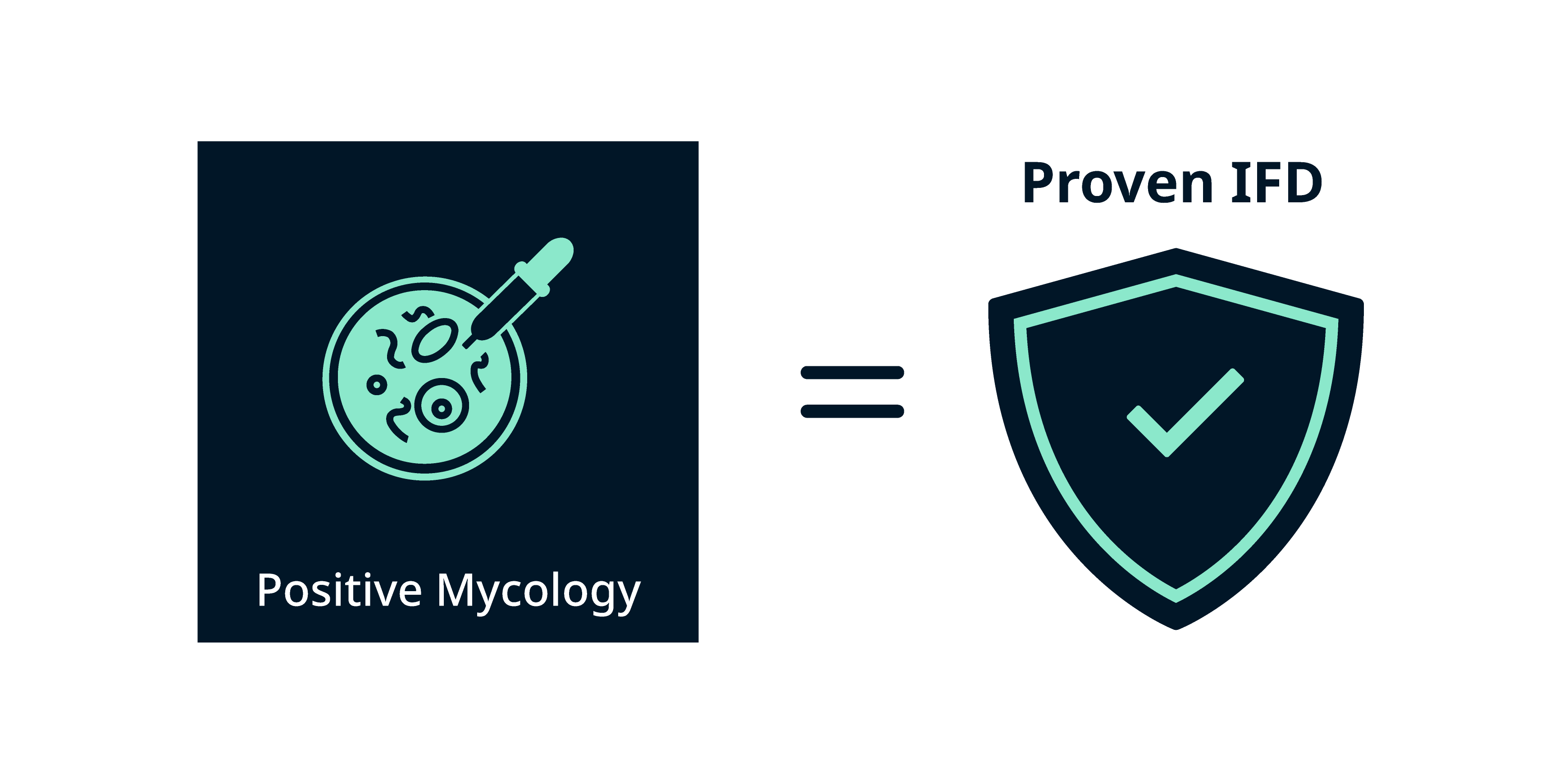
Proven IFD (proposed for any patient, regardless of whether they are immunocompromised) requires:7,10
- Positive mycology: a fungus detected by blood culture or histology/culture of a tissue specimen taken from a normally sterile clinical site
While the EORTC/MSGERC criteria offer comprehensive guidance on the diagnosis of IFDs, their application in practice must be considered in the context of the following points:
- The definitions focus primarily on patients with cancer, and stem cell or solid organ transplant recipients. They may therefore not be suitable for patients outside these categories, especially critically ill patients admitted in the ICU who may have other risk factors for IFDs10
- While the definition of proven IFD applies to a broad range of hosts, the categories of probable and possible IFD were primarily designed for classical immunocompromised hosts, and therefore may not be ideal for other patient populations10
- When considering IFD diagnosis in other populations, particularly critically ill patients, additional definitions have been developed as guidance8,9,12–14
There are multiple considerations
when selecting a diagnostic
test for IFDs:
- The interplay between the host, the microbe and the laboratory setting may affect diagnostic test performance1
- A logical first step is to use serum biomarkers and/or bronchoalveolar lavage specimens for fungal staining, culture and antigen detection1
- A range of tools are currently available to aid clinicians in reaching a diagnosis1
When choosing a diagnostic test (or tests), healthcare professionals should consider the following:2
Sensitivity
The ability of a test or instrument to yield a positive result for a patient with that disease
Specificity
The ability of the test or instrument to obtain a negative result for an individual without that disease
Positive predictive value
How many positive results are ‘true’ out of all positive findings
Negative predictive value
How many negative results are ‘true’ out of all negative findings
Several diagnostic approaches are available for IFDs:
Conventional testing
For many decades, the diagnosis of fungal infections has relied predominantly on methods such as culture testing, direct microscopy and histopathology.1
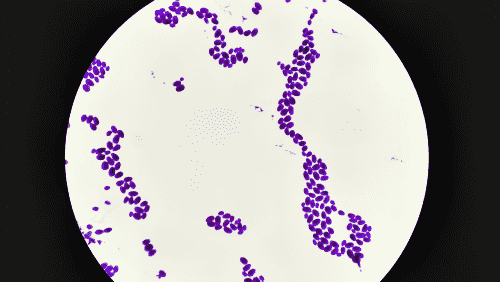 | Culture testing for IFDs, the historic ‘gold standard’, relies on propagating fungal cultures from patient samples such as blood, bronchoalveolar lavage (BAL) fluid or sputum.2,3 The causative pathological strains (such as Candida, Cryptococcus or Fusarium spp.) are subsequently identified through macroscopic or microscopic means.2,3 Specific guidelines should be consulted for specimen-specific considerations. For example, the diagnosis of intra-abdominal candidiasis is often hampered by the lack of specific clinical signs and symptoms, and blood cultures are often negative.4 Samples for diagnosis should be taken intraoperatively, and/or from abdominal drains that have been in place no longer than 24 hours.4 Matrix-assisted laser desorption ionisation–time of flight mass spectrometry (MALDI-TOF MS) is frequently used across Europe for rapid and accessible identification of pathological strains from cultures using databases, reducing the need for highly trained mycology experts.2,5 |
Benefits
- This method is recommended by clinical guidelines, is supported by years of clinical and research experience, and provides the ability to use a range of different sample types3,6
Limitations
- Relatively low sensitivity, depending on the type of fungi (50–95% for yeasts and 1–5% for moulds)7
- Long turnaround times (up to 5 days for yeasts and up to 4 weeks for moulds)2,7
- Relies on mycology experts to interpret results2,7
- Sample contamination can make cultures challenging to interpret8
Future perspectives to develop this method include
- Improved use of surveillance fungal cultures in asymptomatic patients at risk of developing an IFD2
| The use of direct microscopy on clinical specimens and fungal-specific histological stains play an important role in the diagnosis of fungal infections such as Cryptococcus neoformans, Pneumocystis jirovecci, Candida spp., Aspergillus spp., Histoplasma capsulatum, Blastomyces dermatitidis, Coccidioides immitis, Paracoccidioides brasiliensis, and the Mucorales.1 |
Benefits
- This method is supported by years of clinical and research experience, and the ability to identify a variety of fungal pathogens1
Limitations
- Sensitivity of microscopy varies with the individual agent, the source and quality of the specimen, and the skills and experience of the laboratorian1
- May require biopsy of deep tissues, which poses a risk to those patients who are most susceptible to invasive disease1
- Identification based on morphology alone is not always possible, and molecular methods may also need to be employed2
Non-conventional testing
In contrast to conventional testing, these methods rely on detecting analytes, such as nucleic acids, antigens, antibodies and metabolites, to identify fungal pathogens.1–3 These techniques can be performed in addition to conventional testing to improve accuracy in diagnosis.1,4
These methods include:
BDG is a fungal antigen that can be used as both a screening biomarker and a single-use diagnostic biomarker for patients with suspected invasive fungal infection.2 It can also be used as a biomarker for treatment stratification.5 An in vitro diagnostic test has been authorised by the US Food and Drug Administration (FDA) for BDG detection in serum samples,2 and is recommended by European guidelines in the diagnosis of probable IFDs in appropriate clinical settings.1,6
Fungal species
- Serological testing of BDG enables positive detection of many infectious strains, such as Aspergillus spp., Candida spp., Fusarium spp. and Pneumocystis jirovecii2
- This method cannot be used to detect strains such as Cryptococcus spp. or Mucorales2
Benefits
- Good specificity, sensitivity and negative predictive value7,8
- It can be used in the diagnosis of several IFDs, including invasive candidiasis and Pneumocystis jirovecii pneumonia1
Limitations
- Cannot distinguish between different BDG-containing fungal strains7
- Elevated BDG titres in discordance with other host and clinical factors may indicate a false-positive result, which could be the result of conditions that cause translocation of fungal cell wall components9
Mannan is a cell wall component of Candida species that which circulates in the blood during infection.10 Enzyme-linked immunosorbent assays for the detection of mannan have been developed (Platelia™ Candida).10
Fungal species
-
Detects Candida species only10
Benefits
-
It is a non-invasive test, which may be beneficial in high-risk patients10
Limitations
-
Low-to-moderate sensitivity and specificity10
-
Varied sensitivity and specificity across Candida species10
-
No assays are currently authorised by the FDA for the diagnosis of invasive candidiasis11
The polysaccharide capsule of Cryptococcus spp. contains glucuronoxylomannan, an antigen that can be tested for in cerebrospinal fluid (CSF) and serum using latex agglutination (LAT) or enzyme immunoassay (EIA) tests.3 A point-of-care lateral flow assay (LFA) has been developed by IMMY, Inc. that can detect cryptococcal antigen in CSF, serum and urine.3
Cryptococcal antigen testing is the primary method used for screening people living with HIV. It can also be used as an adjunct to culture testing in the diagnosis of cryptococcosis since cultures can be slow growing – for instance, CSF cultures take 3–7 days to grow.3,12
Cryptococcal antigen testing is recommended by the EORTC and the MSGERC as mycological evidence of proven cryptococcosis.1
Fungal species
- Cryptococcus species3
Benefits (LAT/EIA)
- High sensitivity and specificity3
- Can be performed on CSF, serum or plasma samples12
- Provides quantitative results that are predictive of survival and can therefore guide disease management13
Benefits (LFA)
- High sensitivity and specificity12
- CSF and whole blood can be applied to the device directly13
- Can be used in other sample types, including BAL fluid and urine; however, the performance of the test has not been as rigorously assessed in these sample types12
- Low cost3
- Easy to use3
Limitations (LAT/EIA)
- LAT/EIA tests require more elaborate sample preparation compared with LFA13
- The laboratory equipment required impedes the use of the test in a clinical setting13
Limitations (LFA)
- Results can be subject to individual interpretation14
- Does not provide fully quantitative results14
Galactomannan is a cell wall component of Aspergillus spp., Histoplasma capsulatum and Fusarium spp. and can be detected using an ELISA or LFA. For the detection of Aspergillus, the Platelia™ Aspergillus ELISA (Bio-Rad Laboratories) and the sōna Aspergillus galactomannan LFA (IMMY Inc.) have been developed.3,7,15,16 Both are Communauté Européenne (CE) marked and the Platelia™ assay is authorised by the FDA; authorisation is ongoing for the LFA.17,18
Fungal species
- Aspergillus species7
Benefits (ELISA)
- Can be used in BAL fluid, serum, CSF and pleural fluid3
- Demonstrated high sensitivity when performed in BAL fluid, which is highest in patients with neutropenia (100% in patients with neutropenia vs 94.7% in patients without neutropenia)19
- When performed using serum samples in neutropenic patients, sensitivity is high (90%), but is significantly lower in non-neutropenic patients (36.8%; P=0.008)19
Benefits (LFA)
- Good overall performance in BAL fluid, comparable to that of the ELISA test16
- 45-minute turnaround time20
- Can be performed in laboratories with basic equipment21
- Can provide quantitative results (when using an automated reader)17
Limitations (ELISA)
- Antifungal treatment has been shown to reduce the sensitivity of the test3
- The sensitivity of the test is highest among those who have haematological malignancies or who have undergone haematopoietic stem cell transplantation and is lower among those who have undergone solid organ transplant or who are immunocompetant3
- False-positive results may occur as a consequence of cross-reactivity of Fusarium spp., which is thought to be due to interference from Fusarium exoantigens22
- Turnaround times are dependent on the distance from the clinical setting to the laboratory the test is performed in16
- Not all laboratories are able to perform the test16
Limitations (LFA)
- Possible false-positive results due to cross-reactivity with Fusarium spp. and other rare moulds23
- Not fully point-of-care, as samples require pre-treatment, heating and centrifugation17
PCR assays detect the presence of DNA through fungal primers, followed by DNA amplification and sequencing/real-time melting curve analysis; they can be used with a range of sample types, such as tissue, BAL fluid and serum.4 Pan-fungal PCR (which captures ‘all fungi’) and strain-specific PCR can be used to detect fungal pathogens.4
Fungal species
- Several assays are commercially available for the diagnosis of a range of IFDs, including specific assays for Mucorales, Pneumocystis jirovecii, Candida spp., and Aspergillus spp.4
- Assays specific to other species, such as Scedosporium spp., Cryptococcus spp. and Fusarium spp., have also been developed3
Benefits
- PCR assays demonstrate good accuracy and specificity4
- Real-time PCR is a fast-growing area within this technology, with demonstrated high sensitivity4
Limitations
- While PCR assays may theoretically be applied to all fungal species, some organisms are more problematic than others; moulds in particular have strong cell walls that can be difficult to lyse for DNA extraction3
- While sensitivity is generally high, this is variable and depends on the sample type.3,4 For example, serum PCR testing for Aspergillus has shown insufficient performance in patients that have received antifungal prophylaxis24
- Some PCR assay reagents are susceptible to fungal contamination, which can lead to false-positive results3,25
- Traditional PCR methods are not able to quantify amplified DNA, so it is not possible to estimate the microbial burden in the patient’s body. This is of particular importance in IFDs, as it is impossible to determine whether the identified fungal DNA is a result of colonisation or active infection3
T2 magnetic resonance
For Candida species, direct detection assays have been developed for the diagnosis of superficial infections and invasive candidiasis.2
- An FDA-authorised assay combines species-specific PCR with T2 magnetic resonance, with higher sensitivity and specificity compared with blood culture2
- Although the negative predictive value of this assay remains consistently high, the positive predictive value varies widely, depending on the expected prevalence in the represented patient population2
- High equipment and reagent costs are a drawback of this method25
Nucleic acid sequence-based amplification (NASBA)
NASBA is a technique similar to PCR, but amplifies mRNA rather than DNA, meaning that it could detect active disease instead of latent or previous infection.3 The possibility of contamination may also be reduced compared with PCR, due to the isothermal nature of the RNA polymerase used and the fact that RNA is less stable than DNA.3
Lateral flow device
A novel lateral flow device has been developed to detect Aspergillus antigens in human serum and BAL fluid.26 The Aspergillus lateral flow device is an immunochromatographic assay that uses JF5, a monoclonal antibody, that binds with great specificity to an extracellular glycoprotein secreted during active growth of Aspergillus.26 The test has good sensitivity and specificity in BAL fluid samples and moderate sensitivity and good specificity in serum samples; the device also generates results within 15 minutes.26
Next-generation sequencing
Next-generation sequencing to detect microbial cell-free DNA in plasma has emerged as a non-invasive diagnostic method for the early detection and diagnosis of fungal infections when other blood biomarkers are negative.27,28 It also allows for detection of novel or rare pathogens that may be missed or misclassified by conventional tests.27,28
Healthcare professionals should consider several factors when interpreting diagnostic results:
Suitable diagnostic samples
Choosing a suitable diagnostic sample, as some are more appropriate than others for the diagnosis of IFDs1,29
Complementary methodologies
Combining complementary diagnostic methodologies, which has been shown to improve diagnostic performance4,7
Refer to guidelines
Referring to available guidelines and consensus statements to provide support in determining the meaning and significance of diagnostic test results1,6,30

Discover patient case studies
Explore and learn about IFDs with our interactive illustrative case studies
References
Methods for diagnosis
- Formanek PE and Dilling DF. Chest. 2019;156:834–842.
- Chen K et al. BMC Infect Dis. 2017;17:159.
- Kidd SE et al. Front Microbiol. 2020;10:2903.
- Terrero-Salcedo D and Powers-Fletcher MV. J Clin Microbiol. 2020;58:e01487–19.
- Sanguinetti M et al. J Antimicrob Chemother. 2019;74:ii27–ii37.
- Schelenz S et al. Lancet Infect Dis. 2015;15:461–474.
- Donnelly JP et al. Clin Infect Dis. 2020;71:1367–1376.
- Martin-Loeches I et al. Intensive Care Med. 2019;45:789–805.
- Pappas PG et al. Clin Infect Dis. 2016;62:e1–50.
- Bassetti M et al. Clin Infect Dis. 2021;72:S121–S127.
- De Pauw B et al. Clin Infect Dis. 2008;46:1813–1821.
- Schauwvlieghe AFAD et al. Lancet Respir Med. 2018;6:782–792.
- Verweij PE et al. Intensive Care Med. 2020;46:1524–1535.
- Koelher P et al. Lancet Infect Dis. 2021;21:e149–e162.
Considerations when selecting a diagnostic test
- Lass-Flörl C, Samardzic E and Knoll M. Clin Microbiol Infect. 2021;27:1230–1241.
- Faculty of Public Health, Health Knowledge. Available at: https://www.healthknowledge.org.uk/public-health-textbook/disease-causation-diagnostic/2c-diagnosis-screening/statistical-aspects-screening. Accessed July 2023.
Conventional testing
- Kozel TR, Wickes B. Fungal diagnostics. Cold Spring Harbor perspectives in medicine. 2014;4:a019299.
- Terrero-Salcedo D and Powers-Fletcher MV. J Clin Microbiol. 2020;58:e01487-19.
- Donnelly JP et al. Clin Infect Dis. 2020;71:1367–1376.
- Pappas PG et al. Clin Infect Dis. 2016;62:e1–50.
- Salmanton-García J et al. Lancet Microbe. 2023;4:E47–E56.
- Cuenca-Estrella M et al. Clin Microbiol Infect. 2012;18:9–18.
- Fang W et al. J Biomed Sci. 2023;30:42.
- Hall KK and Lyman JA. Clin Microbiol Rev. 2006;19:788–802.
Non-conventional testing and interpretation of results
- Donnelly JP et al. Clin Infect Dis. 2020;71:1367–1376.
- Terrero-Salcedo D and Powers-Fletcher MV. J Clin Microbiol. 2020;58:e01487-19.
- Arvanitis M et al. Clin Microbiol Rev. 2014;27:490–526.
- Kidd SE et al. Front Microbiol. 2020;10:2903.
- Posteraro B et al. J Antimicrob Chemother.2016;71:2262–2269.
- Cuenca-Estrella M et al. Clin Microbiol Infect. 2012;18:9–18.
- Formanek PE and Dilling DF. Chest. 2019;156:834–842.
- Dupuis C et al. Open Forum Infect Dis. 2021;8:ofab080.
- Finkelman MA. J Fungi. 2020;7:14.
- Mikulska M et al. Crit Care. 2010;14:R222.
- Barantsevich N and Barantsevich E. Antibiotics (Basel). 2022;11:718.
- Chang CC et al. Lancet Infect Dis. 2021;21:e375–86.
- Schub T et al. Curr Microbiol. 2021;78:3989–95.
- Huang HR et al. PLoS One. 2015;10:e0127117.
- Sanguinetti M et al. J Antimicrob Chemother. 2019;74:ii27–ii37.
- Jenks JD et al. Clin Infect Dis. 2021;73:e1737–1744.
- Jenks JD, Hoenigl M. Expert Rev Mol Diagn. 2020;20:1009–1017.
- U.S Food and Drug Administration. NOM Product Classification. Available at: https://www.accessdata.fda.gov/scripts/cdrh/cfdocs/cfPCD/classification.cfm?ID=NOM. Accessed July 2023.
- Maertens J et al. Clin Infect Dis. 2010;50:1071–1073.
- Aspergillus GM Lateral Flow Assay. Available at: https://www.bmgrp.eu/uploads/pdf/Aspergillus-Galactomannan-Lateral-Flow-IMMY.pdf. Accessed July 2023.
- Jenks JD et al. Curr Fungal Infect Rep. 2020;14:378–383.
- Tortorano AM et al. J Clin Microbiol. 2012;50:1051–1053.
- Lass-Flörl C et al. Clin Microbiol Infect. 2019;25:1563.
- Egger M et al. J Fungi. 2020;6:18.
- Wickes BL and Wiederhold NP. Nat Commun. 2018;9:5135.
- Pan Z et al. J Med Microbiol. 2015;64:702–707.
- Hoenigl M et al. J Clin Microbiol. 2023;61.3:e01859-22.
- Hong DK et al. Diagn Microbiol Infect Dis. 2018;92:210–213.
- Bassetti M et al. Clin Infect Dis. 2021;72:S121–S127.
- Schelenz S et al. Lancet Infect Dis. 2015;15:461–474.